Settings file¶
Default settings code
1function [stg] = default_settings() 2 3%% Import 4 5% True or false to decide whether to run import functions (Import) 6stg.import = true; 7 8 9% Name of the excel file with the sbtab (SBtab excel name) 10stg.sbtab_excel_name = "SBTAB.xlsx"; 11 12% Name of the model (Name) 13stg.name = "model_name"; 14 15% Name of the default model compartment (Compartment name) 16stg.cname = "Compartment"; 17 18% Name of the sbtab saved in .mat format (SBtab name) 19stg.sbtab_name = "SBtab_" + stg.name; 20 21%% Analysis 22 23% Experiments to run (example experiment 1 to 3 and experimet 6) 24stg.exprun = [1:3,6]; 25 26% Choice between 0,1,2 and 3,4 to choose the scoring method (check 27% documentation) (Use logarithm) 28stg.useLog = 4; 29 30% True or false to decide whether to use multicore everywhere it is 31% available (Optimization Multicore) 32stg.optmc = true; 33 34% Choice of ramdom seed (Ramdom seed) 35stg.rseed = 1; 36 37% True or false to decide whether to use display simulation diagnostics in 38% the console (Simulation Console) 39stg.simcsl = false; 40 41% True or false to decide whether to display optimization results on 42% console (Optimization console) 43stg.optcsl = false; 44 45% True or false to decide whether to save results (Save results) 46stg.save_results = true; 47 48% True or false to decide whether to run detailed simulation for plots 49stg.simdetail = true; 50 51%% Simulation 52 53% Maximum time for each individual function to run in seconds (Maximum 54% time) 55stg.maxt = 10; 56 57% Equilibration time (Equilibration time) 58stg.eqt = 50000; 59 60% True or false to decide whether to do Dimensional Analysis (Dimensional 61% Analysis) 62stg.dimenanal = false; 63 64% True or false to decide whether to do Unit conversion (Unit conversion) 65stg.UnitConversion = false; 66 67% True or false to decide whether to do Absolute Tolerance Scaling 68% (Absolute Tolerance Scaling) 69stg.abstolscale = true; 70 71% Value of Relative tolerance (Relative tolerance) 72stg.reltol = 1.0E-4; 73 74% Value of Absolute tolerance (Absolute tolerance) 75stg.abstol = 1.0E-4; 76 77% Time units for simulation (Simulation time) 78stg.simtime = "second"; 79 80% True or false to decide whether to run sbioaccelerate (after changing 81% this value you need to run "clear functions" to see an effect) 82% (sbioaccelerate) 83stg.sbioacc = true; 84 85% (Absolute tolerance step size for equilibration) 86stg.abstolstepsize_eq = []; 87 88% Max step size in the simulation (if empty matlab decides whats best) 89% (Maximum step) 90stg.maxstep = [10]; 91 92% Max step size in the equilibration (if empty matlab decides whats best) 93% (Maximum step) 94stg.maxstepeq = [2]; 95 96% Max step size in the detailed plots (if empty matlab decides whats best) 97% (Maximum step) 98stg.maxstepdetail = [2]; 99 100% Default score when there is a simulation error, this is needed to keep 101% the optimizations working. 102% (error score) 103stg.errorscore = 10^5; 104%% Model 105 106% Number of parameters to optimize (Parameter number) 107stg.parnum = 5; 108 109original_parameter_set = zeros(1,10); 110 111% Array with the lower bound of all parameters (Lower bound) 112stg.lb = original_parameter_set-5; 113 114% Array with the upper bound of all parameters (Upper bound) 115stg.ub = original_parameter_set+5; 116 117%% Diagnostics 118 119% Choice of what parameters in the array to test, the indices correspond to 120% the parameters in the model and the numbers correspond to the parameters 121% in the optimization array, usually not all parameters are optimized so 122% there needs to be a match between one and the other. (Parameters to test) 123% In this example there are ten parameters in this imaginary model and we 124% are only interested in parameter 2,4,8,9, and 10. Note that stg.parnum is 125% five because of this and not ten 126stg.partest(:,1) = [0,1,0,2,0,0,0,3,4,5]; 127 128% (Parameter array to test) 129stg.pat = [1:2]; 130 131% All the parameter arrays, in this case there is only one (parameters here 132% are in log10 space)(Parameter arrays) 133stg.pa(1,:) = [1,1,1,1,1]; 134stg.pa(1,:) = [1,0,1,2,1]; 135 136% Best parameter array found so far for the model (Best parameter array) 137stg.bestpa = stg.pa(1,:); 138 139%% Plots 140 141% True or false to decide whether to plot results (Plots) 142stg.plot = true; 143 144% True or false to decide whether to use long names in the title of the 145% outputs plots in f_plot_outputs.m (Plot outputs long names) 146stg.plotoln = true; 147 148%% Sensitivity analysis 149 150% Number of samples to use in SA (Sensitivity analysis number of samples) 151stg.sansamples = 100; 152 153% True or false to decide whether to subtract the mean before calculating 154% SI and SIT (Sensitivity analysis subtract mean) 155stg.sasubmean = true; 156 157% Choose the way you want to obtain the samples of the parameters for 158% performing the SA; 0 Log uniform distribution truncated at the parameter 159% bounds 1 Log normal distribution with mu as the best value for a 160% parameter and sigma as stg.sasamplesigma truncated at the parameter 161% bounds 2 same as 1 without truncation 3 Log normal distribution centered 162% at the mean of the parameter bounds and sigma as stg.sasamplesigma 163% truncated at the parameter bounds 4 same as 3 without truncation. 164% (Sensitivity analysis sampling mode) 165stg.sasamplemode = 2; 166 167% Sigma for creating the normal distribution of parameters to perform 168% sensitivity analysis (Sensitivity analysis sampling sigma) 169stg.sasamplesigma = 0.1; 170 171%% Optimization 172 173% Time for the optimization in seconds (fmincon does not respect this 174% time!!) (Optimization time) 175stg.optt = 60*5; 176 177% Population size for the algorithms that use populations (Population size) 178stg.popsize = 144; 179 180% optimization start method, choose between: 1 Random starting point or 181% group of starting points inside the bounds 2 Random starting point or 182% group of starting points near the best point (Optimization start method) 183stg.osm = 1; 184 185% Distance from best parameter array to be used in stg.osm method 2 186% (Distance from best parameter array) 187stg.dbs = 0.1; 188 189% True or false to decide whether to use Multistart (Multistart) 190stg.mst = false; 191 192% Multistart size 193stg.msts = 1; 194 195% True or false to decide whether to display Plots (Plots doesn't work if 196% using multicore) (Optimization plots) 197stg.optplots = true; 198 199% True or false to decide whether to run fmincon (no gradient so this 200% doesn't work very well, no max time!!) 201stg.fmincon = false; 202 203% Options for fmincon (fmincon options) 204stg.fm_options = optimoptions('fmincon',... 205 'Algorithm','interior-point',... 206 'MaxIterations',2,'OptimalityTolerance',0,... 207 'StepTolerance',1e-6,'FiniteDifferenceType','central',... 208 'MaxFunctionEvaluations',10000); 209 210% True or false to decide whether to run simulated annealing (Simulated 211% annealing) 212stg.sa = false; 213 214% Options for simulated annealing (Simulated annealing options) 215stg.sa_options = optimoptions(@simulannealbnd, ... 216 'MaxTime',stg.optt,... 217 'ReannealInterval',40); 218 219% True or false to decide whether to run Pattern search (Pattern search) 220stg.psearch = false; 221 222% Options for Pattern search (Pattern search options) 223stg.psearch_options = optimoptions(@patternsearch,... 224 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 225 'UseCompletePoll',true,'UseCompleteSearch',true,... 226 'MaxMeshSize',2,'MaxFunctionEvaluations',2000); 227 228% True or false to decide whether to run Genetic algorithm (Genetic 229% algorithm) 230stg.ga = false; 231 232% Options for Genetic algorithm (Genetic algorithm options) 233stg.ga_options = optimoptions(@ga,'MaxGenerations',200,... 234 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 235 'PopulationSize',stg.popsize,... 236 'MutationFcn','mutationadaptfeasible'); 237 238% True or false to decide whether to run Particle swarm (Particle swarm) 239stg.pswarm = false; 240 241% Options for Particle swarm (Particle swarm options) 242stg.pswarm_options = optimoptions('particleswarm',... 243 'MaxTime',stg.optt,'UseParallel',stg.optmc,'MaxIterations',200,... 244 'SwarmSize',stg.popsize); 245 246% True or false to decide whether to run Surrogate optimization (Surrogate 247% optimization) 248stg.sopt = false; 249 250% Options for Surrogate optimization (Surrogate optimization options) 251stg.sopt_options = optimoptions('surrogateopt',... 252 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 253 'MaxFunctionEvaluations',5000,... 254 'MinSampleDistance',0.2,'MinSurrogatePoints',32*2+1); 255endExample settings code
1function [stg] = Example_model() 2 3%% Import 4 5% True or false to decide whether to run import functions (Import) 6stg.import = true; 7 8% Name of the excel file with the sbtab (SBtab excel name) 9stg.sbtab_excel_name = "SBTAB example.xlsx"; 10 11% Name of the model (Name) 12stg.name = "Example"; 13 14% Name of the default model compartment (Compartment name) 15stg.cname = "Compartment"; 16 17%% Analysis 18 19% Experiments to run 20stg.exprun = [1,3]; 21% stg.exprun = [1,2,3]; 22 23% Choice between 0,1,2 and 3 to change either and how to apply log10 to the 24% scores (check documentation) (Use logarithm) 25stg.useLog = 0; 26 27% True or false to decide whether to use multicore everywhere it is 28% available (Optimization Multicore) 29stg.optmc = false; 30 31% Choice of ramdom seed (Ramdom seed) 32stg.rseed = 1; 33 34% True or false to decide whether to use display simulation diagnostics in 35% the console (Simulation Console) 36stg.simcsl = false; 37 38% True or false to decide whether to display optimization results on 39% console (Optimization console) 40stg.optcsl = true; 41 42% True or false to decide whether to display PLA results on console (PLA 43% console) 44stg.placsl = true; 45 46% True or false to decide whether to save results (Save results) 47stg.save_results = true; 48 49% True or false to decide whether to run detailed simulation for plots 50stg.simdetail = false; 51 52%% Simulation 53 54% Maximum time for each individual function to run in seconds (Maximum 55% time) 56stg.maxt = 2; 57 58% Equilibration time (Equilibration time) 59stg.eqt = 50000; 60 61% True or false to decide whether to do Dimensional Analysis (Dimensional 62% Analysis) 63stg.dimenanal = true; 64 65% True or false to decide whether to do Unit conversion (Unit conversion) 66stg.UnitConversion = true; 67 68% True or false to decide whether to do Absolute Tolerance Scaling 69% (Absolute Tolerance Scaling) 70stg.abstolscale = true; 71 72% Value of Relative tolerance (Relative tolerance) 73stg.reltol = 1.0E-4; 74 75% Value of Absolute tolerance (Absolute tolerance) 76stg.abstol = 1.0E-7; 77 78% Time units for simulation (Simulation time) 79stg.simtime = "second"; 80 81% True or false to decide whether to run sbioaccelerate (after changing 82% this value you need to run "clear functions" to see an effect) 83% (sbioaccelerate) 84stg.sbioacc = false; 85 86% Max step size in the simulation (if empty matlab decides whats best) 87% (Maximum step) 88stg.maxstep = []; 89 90% Max step size in the equilibration (if empty matlab decides whats best) 91% (Maximum step) 92stg.maxstepeq = []; 93 94% Max step size in the detailed plots (if empty matlab decides whats best) 95% (Maximum step) 96stg.maxstepdetail = [0.001]; 97 98% Default score when there is a simulation error, this is needed to keep 99% the optimizations working. (error score) 100stg.errorscore = 10^5; 101 102%% Model 103 104% Number of parameters to optimize (Parameter number) 105stg.parnum = 12; 106 107% Index for the parameters that have thermodynamic constrains (Termodiamic 108% Constrains Index) 109stg.tci = [8]; 110 111% Parameters to multiply to the first parameter (in Stg.partest to get to 112% the correct thermodynamic constrain formula) (Termodiamic Constrains 113% multipliers) 114stg.tcm([8],1) = [4]; 115stg.tcm([8],2) = [5]; 116stg.tcm([8],3) = [7]; 117 118% Parameters to divide to the first parameter (in Stg.partest to get to the 119% correct thermodynamic constrain formula) (Termodiamic Constrains 120% divisors) 121stg.tcd([8],1) = [1]; 122stg.tcd([8],2) = [3]; 123stg.tcd([8],3) = [6]; 124 125% Array with the lower bound of all parameters (Lower bound) 126stg.lb = zeros(1,stg.parnum)-4; 127 128% Array with the upper bound of all parameters (Upper bound) 129stg.ub = zeros(1,stg.parnum)+4; 130 131%% Diagnostics 132 133% Choice of what parameters in the array to test, the indices correspond to 134% the parameters in the model and the numbers correspond to the parameters 135% in the optimization array, usually not all parameters are optimized so 136% there needs to be a match between one and the other. (Parameters to test) 137stg.partest(:,1) = [1 ,2 ,3 ,4 ,5 ,6 ,7 ,2 ,8 ,9,... 138 10 ,11 ,12]; 139 140% (Parameter array to test) 141stg.pat = 1:3; 142 143% All the parameter arrays, in this case there is only one (Parameter 144% arrays) 145stg.pa(1,:) = [3.999,0.696,1.072,3.429,-0.751,-3.741,-0.569,0.831,3.068,0.921,-2.156,-1.970]; 146stg.pa(2,:) = stg.pa(1,:)-1; 147stg.pa(3,:) = stg.pa(1,:)+1; 148 149% Best parameter array found so far for the model (Best parameter array) 150stg.bestpa = stg.pa(1,:); 151 152%% Plots 153 154% True or false to decide whether to plot results (Plots) 155stg.plot = true; 156 157% True or false to decide whether to use long names in the title of the 158% outputs plots in f_plot_outputs.m (Plot outputs long names) 159stg.plotnames = true; 160 161%% Sensitivity analysis 162 163% Number of samples to use in SA (Sensitivity analysis number of samples) 164stg.sansamples = 1000; 165 166% True or false to decide whether to subtract the mean before calculating 167% SI and SIT (Sensitivity analysis subtract mean) 168stg.sasubmean = true; 169 170% Choose the way you want to obtain the samples of the parameters for 171% performing the SA; 0 Log uniform distribution truncated at the parameter 172% bounds 1 Log normal distribution with mu as the best value for a 173% parameter and sigma as stg.sasamplesigma truncated at the parameter 174% bounds 2 same as 1 without truncation 3 Log normal distribution centered 175% at the mean of the parameter bounds and sigma as stg.sasamplesigma 176% truncated at the parameter bounds 4 same as 3 without truncation. 177% (Sensitivity analysis sampling mode) 178stg.sasamplemode = 2; 179 180% Sigma for creating the normal distribution of parameters to perform 181% sensitivity analysis (Sensitivity analysis sampling sigma) 182stg.sasamplesigma = 0.1; 183 184stg.gsabootstrapsize = ceil(sqrt(stg.sansamples)); 185 186%% Optimization 187 188% Time for the optimization in seconds (fmincon does not respect this 189% time!!) (Optimization time) 190stg.optt = 60*1; 191 192% Population size for the algorithms that use populations (Population size) 193stg.popsize = 10; 194 195% optimization start method, choose between: 1 Random starting point or 196% group of starting points inside the bounds 2 Random starting point or 197% group of starting points near the best point (Optimization start method) 198stg.osm = 1; 199 200% Distance from best parameter array to be used in stg.osm method 2 201% (Distance from best parameter array) 202stg.dbs = 0.1; 203 204% True or false to decide whether to use Multistart (Multistart) 205stg.mst = false; 206 207% Multistart size 208stg.msts = 1; 209 210% True or false to decide whether to display Plots (Plots doesn't work if 211% using multicore) (Optimization plots) 212stg.optplots = true; 213 214% True or false to decide whether to run fmincon (no gradient so this 215% doesn't work very well, no max time!!) 216stg.fmincon = false; 217 218% Options for fmincon (fmincon options) 219stg.fm_options = optimoptions('fmincon',... 220 'UseParallel',stg.optmc,... 221 'Algorithm','interior-point',... 222 'MaxIterations',2,'OptimalityTolerance',0,... 223 'StepTolerance',1e-6,'FiniteDifferenceType','central',... 224 'MaxFunctionEvaluations',10000); 225 226% True or false to decide whether to run simulated annealing (Simulated 227% annealing) 228stg.sa = false; 229 230% Options for simulated annealing (Simulated annealing options) 231stg.sa_options = optimoptions(@simulannealbnd, ... 232 'MaxTime',stg.optt,... 233 'InitialTemperature',... 234 ones(1,stg.parnum)*2,'ReannealInterval',40); 235 236% True or false to decide whether to run Pattern search (Pattern search) 237stg.psearch = false; 238 239% Options for Pattern search (Pattern search options) 240stg.psearch_options = optimoptions(@patternsearch,... 241 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 242 'UseCompletePoll',true,'UseCompleteSearch',true,... 243 'MaxMeshSize',2,'MaxFunctionEvaluations',2000); 244 245% True or false to decide whether to run Genetic algorithm (Genetic 246% algorithm) 247stg.ga = true; 248 249% Options for Genetic algorithm (Genetic algorithm options) 250stg.ga_options = optimoptions(@ga,'MaxGenerations',200,... 251 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 252 'PopulationSize',stg.popsize,... 253 'MutationFcn','mutationadaptfeasible','Display','diagnose'); 254 255% True or false to decide whether to run Particle swarm (Particle swarm) 256stg.pswarm = false; 257 258% Options for Particle swarm (Particle swarm options) 259stg.pswarm_options = optimoptions('particleswarm',... 260 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 261 'SwarmSize',stg.popsize); 262 263% True or false to decide whether to run Surrogate optimization (Surrogate 264% optimization) 265stg.sopt = false; 266 267% Options for Surrogate optimization (Surrogate optimization options) 268stg.sopt_options = optimoptions('surrogateopt',... 269 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 270 'MaxFunctionEvaluations',5000,... 271 'MinSampleDistance',0.2,'MinSurrogatePoints',32*2+1); 272end
Import¶
Default settings code
1% True or false to decide whether to run import functions (Import) 2stg.import = true; 3 4 5% Name of the excel file with the sbtab (SBtab excel name) 6stg.sbtab_excel_name = "SBTAB.xlsx"; 7 8% Name of the model (Name) 9stg.name = "model_name"; 10 11% Name of the default model compartment (Compartment name) 12stg.cname = "Compartment"; 13 14% Name of the sbtab saved in .mat format (SBtab name) 15stg.sbtab_name = "SBtab_" + stg.name;Example settings code
1% True or false to decide whether to run import functions (Import) 2stg.import = true; 3 4% Name of the excel file with the sbtab (SBtab excel name) 5stg.sbtab_excel_name = "SBTAB example.xlsx"; 6 7% Name of the model (Name) 8stg.name = "Example"; 9 10% Name of the default model compartment (Compartment name) 11stg.cname = "Compartment";
stg.import - (logical) Decide whether to run import functions
stg.sbtab_excel_name - (string) Name of the Excel file with the SBtab
stg.name - (string) Name of the model
stg.cname - (string) Name of the default model compartment
stg.sbtab_name - (string) Name of the SBtab saved in .mat format
Analysis¶
Default settings code
1% Experiments to run (example experiment 1 to 3 and experimet 6) 2stg.exprun = [1:3,6]; 3 4% Choice between 0,1,2 and 3,4 to choose the scoring method (check 5% documentation) (Use logarithm) 6stg.useLog = 4; 7 8% True or false to decide whether to use multicore everywhere it is 9% available (Optimization Multicore) 10stg.optmc = true; 11 12% Choice of ramdom seed (Ramdom seed) 13stg.rseed = 1; 14 15% True or false to decide whether to use display simulation diagnostics in 16% the console (Simulation Console) 17stg.simcsl = false; 18 19% True or false to decide whether to display optimization results on 20% console (Optimization console) 21stg.optcsl = false; 22 23% True or false to decide whether to save results (Save results) 24stg.save_results = true; 25 26% True or false to decide whether to run detailed simulation for plots 27stg.simdetail = true;Example settings code
1% Experiments to run 2stg.exprun = [1,3]; 3% stg.exprun = [1,2,3]; 4 5% Choice between 0,1,2 and 3 to change either and how to apply log10 to the 6% scores (check documentation) (Use logarithm) 7stg.useLog = 0; 8 9% True or false to decide whether to use multicore everywhere it is 10% available (Optimization Multicore) 11stg.optmc = false; 12 13% Choice of ramdom seed (Ramdom seed) 14stg.rseed = 1; 15 16% True or false to decide whether to use display simulation diagnostics in 17% the console (Simulation Console) 18stg.simcsl = false; 19 20% True or false to decide whether to display optimization results on 21% console (Optimization console) 22stg.optcsl = true; 23 24% True or false to decide whether to display PLA results on console (PLA 25% console) 26stg.placsl = true; 27 28% True or false to decide whether to save results (Save results) 29stg.save_results = true; 30 31% True or false to decide whether to run detailed simulation for plots 32stg.simdetail = false;
stg.exprun - (double) Experiments to run
stg.useLog - (double) Choice between 0,1,2 and 3 to change either and how to apply log10 to the scores, check results:
stg.optmc - (logical) Decide whether to use multicore everywhere it is available
stg.rseed - (double) Choice of random seed
stg.simcsl - (logical) Decide whether to display simulation diagnostics in the console
stg.optcsl - (logical) Decide whether to display optimization results on console
stg.save_results - (logical) Decide whether to save results
stg.simdetail - (logical) Decide whether to run detailed simulation for plots
Simulation¶
Default settings code
1% Maximum time for each individual function to run in seconds (Maximum 2% time) 3stg.maxt = 10; 4 5% Equilibration time (Equilibration time) 6stg.eqt = 50000; 7 8% True or false to decide whether to do Dimensional Analysis (Dimensional 9% Analysis) 10stg.dimenanal = false; 11 12% True or false to decide whether to do Unit conversion (Unit conversion) 13stg.UnitConversion = false; 14 15% True or false to decide whether to do Absolute Tolerance Scaling 16% (Absolute Tolerance Scaling) 17stg.abstolscale = true; 18 19% Value of Relative tolerance (Relative tolerance) 20stg.reltol = 1.0E-4; 21 22% Value of Absolute tolerance (Absolute tolerance) 23stg.abstol = 1.0E-4; 24 25% Time units for simulation (Simulation time) 26stg.simtime = "second"; 27 28% True or false to decide whether to run sbioaccelerate (after changing 29% this value you need to run "clear functions" to see an effect) 30% (sbioaccelerate) 31stg.sbioacc = true; 32 33% (Absolute tolerance step size for equilibration) 34stg.abstolstepsize_eq = []; 35 36% Max step size in the simulation (if empty matlab decides whats best) 37% (Maximum step) 38stg.maxstep = [10]; 39 40% Max step size in the equilibration (if empty matlab decides whats best) 41% (Maximum step) 42stg.maxstepeq = [2]; 43 44% Max step size in the detailed plots (if empty matlab decides whats best) 45% (Maximum step) 46stg.maxstepdetail = [2]; 47 48% Default score when there is a simulation error, this is needed to keep 49% the optimizations working. 50% (error score) 51stg.errorscore = 10^5;Example settings code
1% Maximum time for each individual function to run in seconds (Maximum 2% time) 3stg.maxt = 2; 4 5% Equilibration time (Equilibration time) 6stg.eqt = 50000; 7 8% True or false to decide whether to do Dimensional Analysis (Dimensional 9% Analysis) 10stg.dimenanal = true; 11 12% True or false to decide whether to do Unit conversion (Unit conversion) 13stg.UnitConversion = true; 14 15% True or false to decide whether to do Absolute Tolerance Scaling 16% (Absolute Tolerance Scaling) 17stg.abstolscale = true; 18 19% Value of Relative tolerance (Relative tolerance) 20stg.reltol = 1.0E-4; 21 22% Value of Absolute tolerance (Absolute tolerance) 23stg.abstol = 1.0E-7; 24 25% Time units for simulation (Simulation time) 26stg.simtime = "second"; 27 28% True or false to decide whether to run sbioaccelerate (after changing 29% this value you need to run "clear functions" to see an effect) 30% (sbioaccelerate) 31stg.sbioacc = false; 32 33% Max step size in the simulation (if empty matlab decides whats best) 34% (Maximum step) 35stg.maxstep = []; 36 37% Max step size in the equilibration (if empty matlab decides whats best) 38% (Maximum step) 39stg.maxstepeq = []; 40 41% Max step size in the detailed plots (if empty matlab decides whats best) 42% (Maximum step) 43stg.maxstepdetail = [0.001]; 44 45% Default score when there is a simulation error, this is needed to keep 46% the optimizations working. (error score) 47stg.errorscore = 10^5;
stg.maxt - (double) Maximum time for each individual function has to run in seconds
stg.eqt - (double) Equilibration time in seconds
stg.dimenanal - (logical) Decide whether to do Dimensional Analysis
stg.UnitConversion - (logical) Decide whether to do Unit conversion
stg.abstolscale - (logical) Decide whether to do Absolute Tolerance Scaling
stg.reltol - (double) Value of Relative tolerance
stg.abstol - (double) Value of Absolute tolerance
stg.simtime - (string) Time units for simulation
stg.sbioacc - (logical) Decide whether to run sbioaccelerate (after changing this value you need to run “clear functions” to see an effect)
stg.abstolstepsize_eq - (double) Absolute tolerance step size for equilibration (if empty MATLAB® decides whats best)
stg.maxstep - (double) Max step size in the simulation (if empty MATLAB® decides what’s best)
stg.maxstepeq - (double) Max step size in the equilibration (if empty MATLAB® decides whats best)
stg.maxstepdetail - (double) Max step size in the detailed plots (if empty MATLAB® decides whats best)
stg.errorscore - (double) Default score when there is a simulation error, this is needed to keep the optimizations working.
Model¶
Default settings code
1% Number of parameters to optimize (Parameter number) 2stg.parnum = 5; 3 4original_parameter_set = zeros(1,10); 5 6% Array with the lower bound of all parameters (Lower bound) 7stg.lb = original_parameter_set-5; 8 9% Array with the upper bound of all parameters (Upper bound) 10stg.ub = original_parameter_set+5;Example settings code
1% Number of parameters to optimize (Parameter number) 2stg.parnum = 12; 3 4% Index for the parameters that have thermodynamic constrains (Termodiamic 5% Constrains Index) 6stg.tci = [8]; 7 8% Parameters to multiply to the first parameter (in Stg.partest to get to 9% the correct thermodynamic constrain formula) (Termodiamic Constrains 10% multipliers) 11stg.tcm([8],1) = [4]; 12stg.tcm([8],2) = [5]; 13stg.tcm([8],3) = [7]; 14 15% Parameters to divide to the first parameter (in Stg.partest to get to the 16% correct thermodynamic constrain formula) (Termodiamic Constrains 17% divisors) 18stg.tcd([8],1) = [1]; 19stg.tcd([8],2) = [3]; 20stg.tcd([8],3) = [6]; 21 22% Array with the lower bound of all parameters (Lower bound) 23stg.lb = zeros(1,stg.parnum)-4; 24 25% Array with the upper bound of all parameters (Upper bound) 26stg.ub = zeros(1,stg.parnum)+4;
stg.parnum - (double) Number of parameters to optimize
stg.tci - (double) Index for the parameters that have thermodynamic constraints
stg.tcm - (double) Parameters to multiply to the first parameter (in stg.partest to get to the correct thermodynamic constraint formula)
stg.tcd - (double) Parameters to divide to the first parameter (in stg.partest to get to the correct thermodynamic constraint formula)
stg.lb - (double) Lower bound of all parameters
\[stg.lb = \begin{bmatrix} lb_{1} & lb_{2} & ... & lb_{i} \end{bmatrix}\]\(i =\) Parameter index
stg.ub - (double) Upper bound of all parameters
\[stg.up = \begin{bmatrix} ub_{1} & ub_{2} & ... & ub_{i} \end{bmatrix}\]\(i =\) Parameter index
Diagnostics¶
Default settings code
1% Choice of what parameters in the array to test, the indices correspond to 2% the parameters in the model and the numbers correspond to the parameters 3% in the optimization array, usually not all parameters are optimized so 4% there needs to be a match between one and the other. (Parameters to test) 5% In this example there are ten parameters in this imaginary model and we 6% are only interested in parameter 2,4,8,9, and 10. Note that stg.parnum is 7% five because of this and not ten 8stg.partest(:,1) = [0,1,0,2,0,0,0,3,4,5]; 9 10% (Parameter array to test) 11stg.pat = [1:2]; 12 13% All the parameter arrays, in this case there is only one (parameters here 14% are in log10 space)(Parameter arrays) 15stg.pa(1,:) = [1,1,1,1,1]; 16stg.pa(1,:) = [1,0,1,2,1]; 17 18% Best parameter array found so far for the model (Best parameter array) 19stg.bestpa = stg.pa(1,:);Example settings code
1% Choice of what parameters in the array to test, the indices correspond to 2% the parameters in the model and the numbers correspond to the parameters 3% in the optimization array, usually not all parameters are optimized so 4% there needs to be a match between one and the other. (Parameters to test) 5stg.partest(:,1) = [1 ,2 ,3 ,4 ,5 ,6 ,7 ,2 ,8 ,9,... 6 10 ,11 ,12]; 7 8% (Parameter array to test) 9stg.pat = 1:3; 10 11% All the parameter arrays, in this case there is only one (Parameter 12% arrays) 13stg.pa(1,:) = [3.999,0.696,1.072,3.429,-0.751,-3.741,-0.569,0.831,3.068,0.921,-2.156,-1.970]; 14stg.pa(2,:) = stg.pa(1,:)-1; 15stg.pa(3,:) = stg.pa(1,:)+1; 16 17% Best parameter array found so far for the model (Best parameter array) 18stg.bestpa = stg.pa(1,:);
stg.partest - (double) Choice of which parameters to work on, since depending on the task, not all SBtab parameters are worked on. k indices correspond to the parameters in the SBtab and numbers up to i correspond to the parameters in the work set. This is the set that actually gets used for diagnostics, optimization, and sensitivity analyis.
\[stg.partest_k = \begin{bmatrix} 1_{k_1} & 2_{k_2} & ... & i_{k_{end}} \end{bmatrix}\]In our example model parameter 216 from the SBtab is parameter number 1 of the work set, parameter 217 from the SBtab is parameter number 2 of the work set, and successively.
\[stg.partest_{[216:227]} = \begin{bmatrix} 1_{216} & 2_{217} & ... & 6_{221} & 1_{222} & 2_{223} & ... & 6_{227} \end{bmatrix}\]stg.pat - (double) Index(\(j\)) of the parameter set to work on
stg.pa - (double) All the parameter sets
\[\begin{split}stg.pa = \begin{bmatrix} x_{1,1} & x_{2,1} & ... & x_{i,1} \\ x_{1,2} & x_{2,2} & ... & x_{i,2} \\ ... & ... & ... & ... \\ x_{1,j} & x_{2,j} & ... & x_{i,j} \end{bmatrix}\end{split}\]stg.bestpa - (double) Best parameter set found so far during optimization
\[stg.bestx = \begin{bmatrix} bestx_{1} & bestx_{2} & ... & bestx_{i} \end{bmatrix}\]\(x =\) Parameters being worked on
\(i =\) Index of Parameters being worked on
\(k =\) Index of the parameters in SBtab
\(j =\) Index of the Parameter set to work on
Plots¶
Default settings code
1% True or false to decide whether to plot results (Plots) 2stg.plot = true; 3 4% True or false to decide whether to use long names in the title of the 5% outputs plots in f_plot_outputs.m (Plot outputs long names) 6stg.plotoln = true;Example settings code
1% True or false to decide whether to plot results (Plots) 2stg.plot = true; 3 4% True or false to decide whether to use long names in the title of the 5% outputs plots in f_plot_outputs.m (Plot outputs long names) 6stg.plotnames = true;
stg.plot - (logical) Decide whether to plot results
stg.plotoln - (logical) Decide whether to use long names in the title of the output plots in f_plot_outputs.m
Global Sensitivity Analysis (GSA)¶
Default settings code
1% Number of samples to use in SA (Sensitivity analysis number of samples) 2stg.sansamples = 100; 3 4% True or false to decide whether to subtract the mean before calculating 5% SI and SIT (Sensitivity analysis subtract mean) 6stg.sasubmean = true; 7 8% Choose the way you want to obtain the samples of the parameters for 9% performing the SA; 0 Log uniform distribution truncated at the parameter 10% bounds 1 Log normal distribution with mu as the best value for a 11% parameter and sigma as stg.sasamplesigma truncated at the parameter 12% bounds 2 same as 1 without truncation 3 Log normal distribution centered 13% at the mean of the parameter bounds and sigma as stg.sasamplesigma 14% truncated at the parameter bounds 4 same as 3 without truncation. 15% (Sensitivity analysis sampling mode) 16stg.sasamplemode = 2; 17 18% Sigma for creating the normal distribution of parameters to perform 19% sensitivity analysis (Sensitivity analysis sampling sigma) 20stg.sasamplesigma = 0.1;Example settings code
1% Number of samples to use in SA (Sensitivity analysis number of samples) 2stg.sansamples = 1000; 3 4% True or false to decide whether to subtract the mean before calculating 5% SI and SIT (Sensitivity analysis subtract mean) 6stg.sasubmean = true; 7 8% Choose the way you want to obtain the samples of the parameters for 9% performing the SA; 0 Log uniform distribution truncated at the parameter 10% bounds 1 Log normal distribution with mu as the best value for a 11% parameter and sigma as stg.sasamplesigma truncated at the parameter 12% bounds 2 same as 1 without truncation 3 Log normal distribution centered 13% at the mean of the parameter bounds and sigma as stg.sasamplesigma 14% truncated at the parameter bounds 4 same as 3 without truncation. 15% (Sensitivity analysis sampling mode) 16stg.sasamplemode = 2; 17 18% Sigma for creating the normal distribution of parameters to perform 19% sensitivity analysis (Sensitivity analysis sampling sigma) 20stg.sasamplesigma = 0.1; 21 22stg.gsabootstrapsize = ceil(sqrt(stg.sansamples));
stg.sansamples - (double) Number of samples to use in GSA (in total (2+npars)*sansamples simulations will be performed, where npars are the number of parameters).
stg.sasubmean - (logical) Decide whether to subtract mean before calculating SI and STI, see Halnes et al 2009.
stg.sasamplemode - (double) Choose the way you want to obtain the samples of the parameters for performing the GSA;
Reciprocal (log uniform) distribution
\(X_{i} \sim Reciprocal(a_{i},b_{i})\)
\(i =\) Parameter index
\(a_{i} = stg.lb_{i}\)
\(b_{i} = stg.ub_{i}\)
Example distribution with \(a = -1, b = 1\)
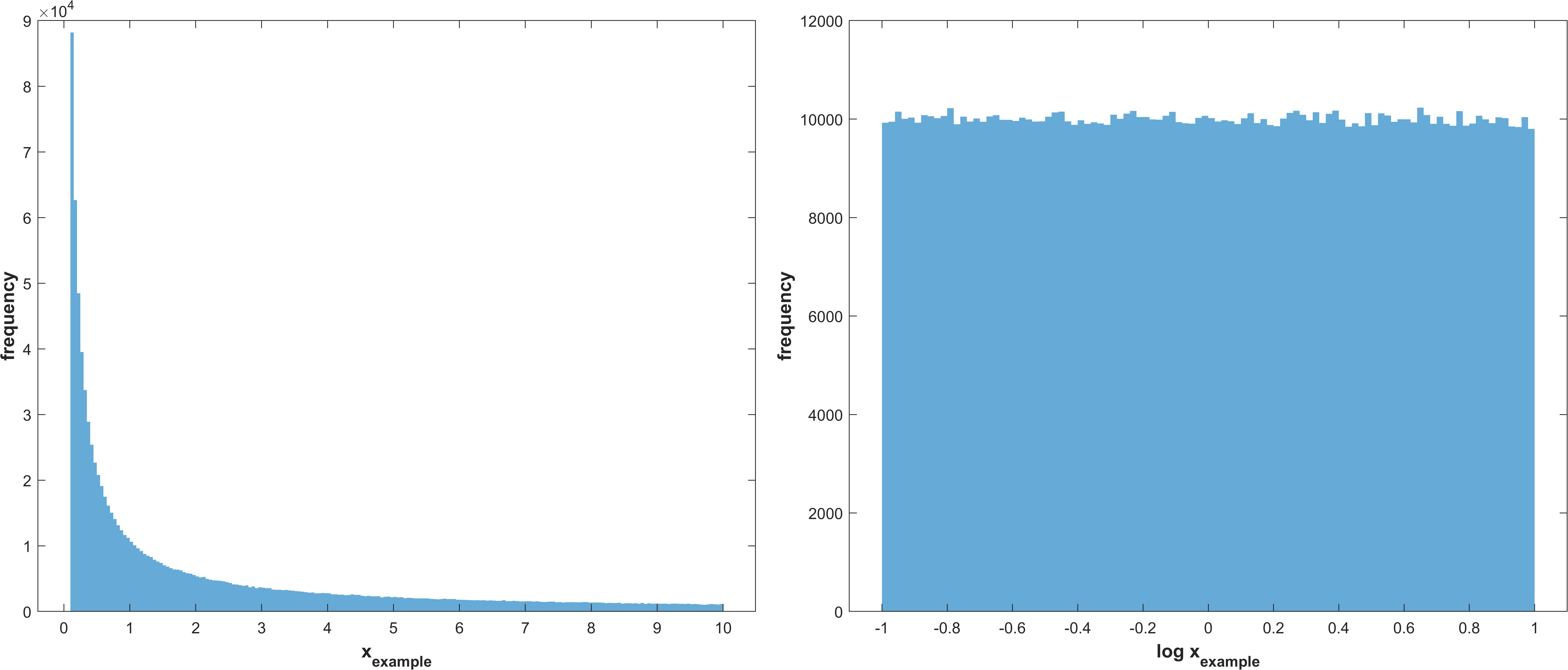
Log normal distribution with μ corresponding to the best value for a parameter, as recieved from the optimization, and σ as stg.sasamplesigma truncated at the parameter bounds
\(X_{i} \sim TruncatedLogNormal(μ_{i}, σ, a_{i}, b_{i})\)
\(i =\) Parameter index
\(μ_{i} = bestx_{i}\)
\(σ = stg.sasamplesigma\)
\(a_{i} = stg.lb_{i}\)
\(b_{i} = stg.ub_{i}\)
Example distribution with \(μ = 0.5, σ = 1, a = -1, b = 1\)
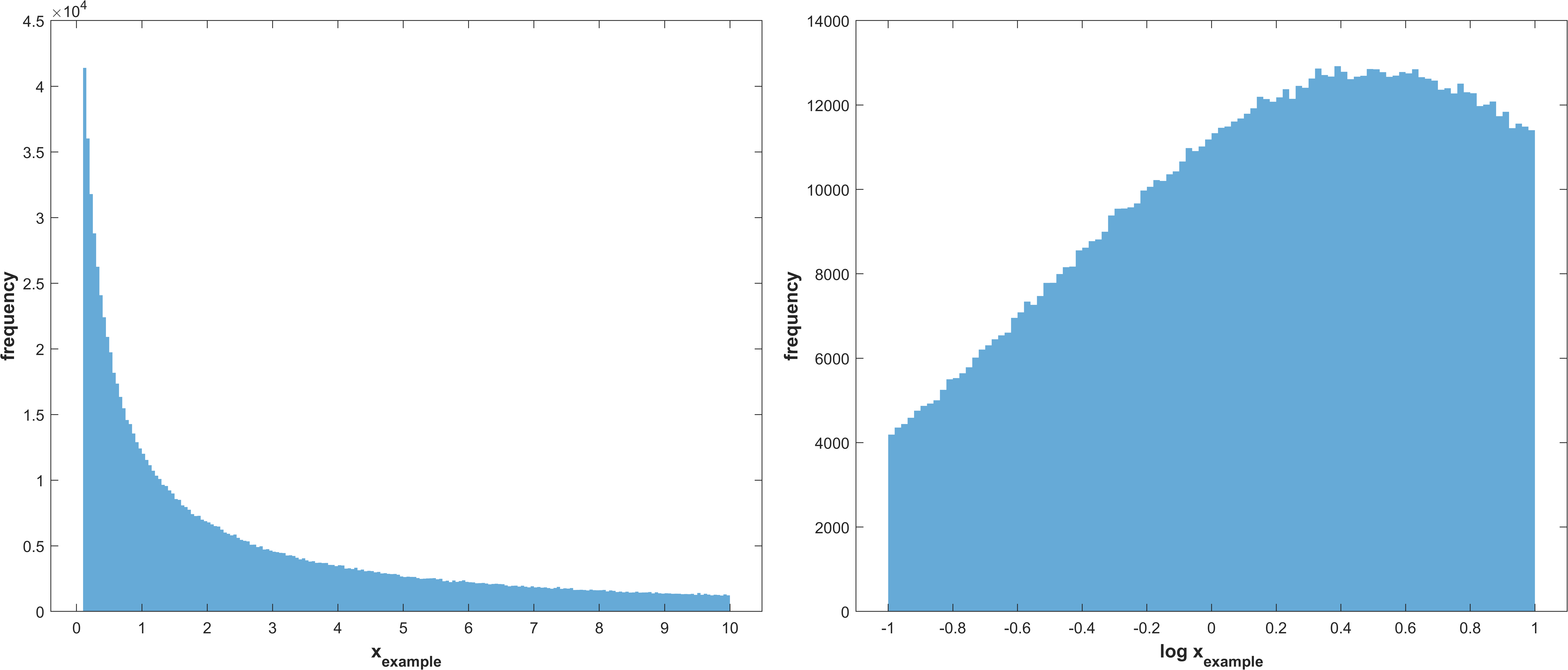
same as 1 without truncation
\(X_{i} \sim LogNormal(μ, σ)\)
\(i =\) Parameter index
\(μ_{i} = bestx_{i}\)
\(σ = stg.sasamplesigma\)
Example distribution with \(μ = 0.5, σ = 1\)
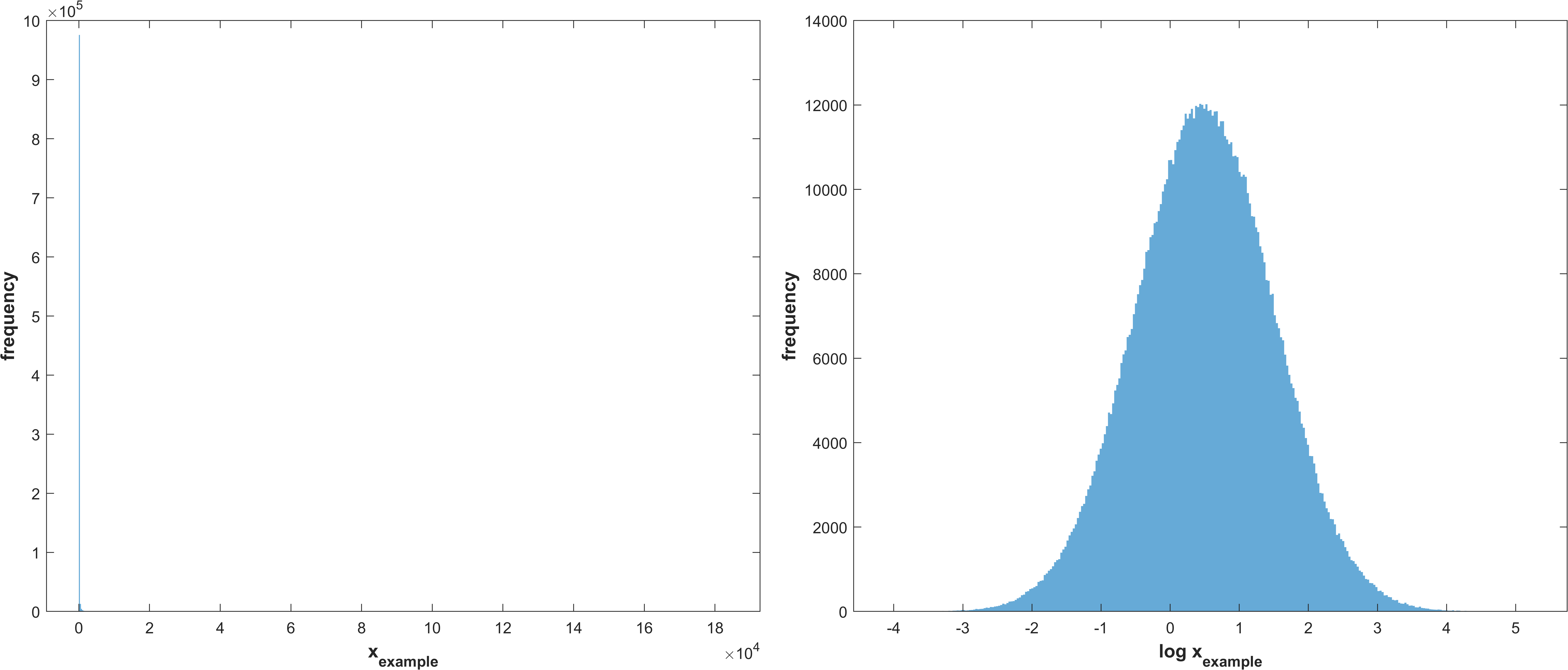
Log normal distribution with μ corresponding to the mean of the parameter bounds and σ as stg.sasamplesigma but truncated at the parameter bounds
\(X_{i} \sim TruncatedLogNormal(μ_{i}, σ, a_{i}, b_{i})\)
\(i =\) Parameter index
\(μ_{i} = \frac{stg.lb_{i} + (stg.ub_{i} - stg.lb_{i})}{2}\)
\(σ = stg.sasamplesigma\)
\(a_{i} = stg.lb_{i}\)
\(b_{i} = stg.ub_{i}\)
Example distribution with \(μ = \frac{a+(b-a)}{2}, σ = 1, a = -1, b = 1\)
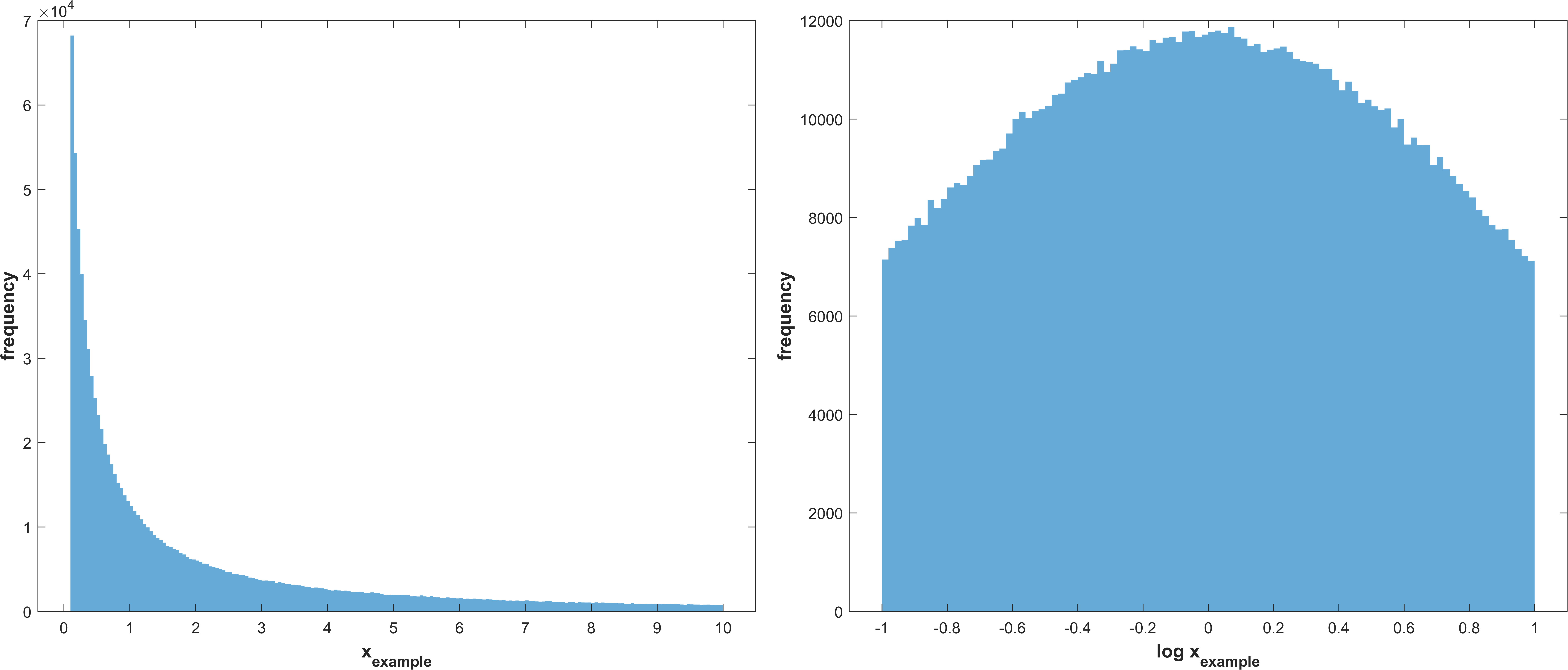
same as 3 without truncation.
\(X_{i} \sim LogNormal(mu_{i}, σ)\)
\(i =\) Parameter index
\(μ_{i} = \frac{stg.lb_{i} + (stg.ub_{i} - stg.lb_{i})}{2}\)
\(σ = stg.sasamplesigma\)
Example distribution with \(μ = \frac{a+(b-a)}{2}, σ = 1, a = -1, b = 1\)
stg.sasamplesigma - (double) σ for creating the normal distribution of parameters to perform sensitivity analysis
Optimization¶
Default settings code
1% Time for the optimization in seconds (fmincon does not respect this 2% time!!) (Optimization time) 3stg.optt = 60*5; 4 5% Population size for the algorithms that use populations (Population size) 6stg.popsize = 144; 7 8% optimization start method, choose between: 1 Random starting point or 9% group of starting points inside the bounds 2 Random starting point or 10% group of starting points near the best point (Optimization start method) 11stg.osm = 1; 12 13% Distance from best parameter array to be used in stg.osm method 2 14% (Distance from best parameter array) 15stg.dbs = 0.1; 16 17% True or false to decide whether to use Multistart (Multistart) 18stg.mst = false; 19 20% Multistart size 21stg.msts = 1; 22 23% True or false to decide whether to display Plots (Plots doesn't work if 24% using multicore) (Optimization plots) 25stg.optplots = true; 26 27% True or false to decide whether to run fmincon (no gradient so this 28% doesn't work very well, no max time!!) 29stg.fmincon = false; 30 31% Options for fmincon (fmincon options) 32stg.fm_options = optimoptions('fmincon',... 33 'Algorithm','interior-point',... 34 'MaxIterations',2,'OptimalityTolerance',0,... 35 'StepTolerance',1e-6,'FiniteDifferenceType','central',... 36 'MaxFunctionEvaluations',10000); 37 38% True or false to decide whether to run simulated annealing (Simulated 39% annealing) 40stg.sa = false; 41 42% Options for simulated annealing (Simulated annealing options) 43stg.sa_options = optimoptions(@simulannealbnd, ... 44 'MaxTime',stg.optt,... 45 'ReannealInterval',40); 46 47% True or false to decide whether to run Pattern search (Pattern search) 48stg.psearch = false; 49 50% Options for Pattern search (Pattern search options) 51stg.psearch_options = optimoptions(@patternsearch,... 52 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 53 'UseCompletePoll',true,'UseCompleteSearch',true,... 54 'MaxMeshSize',2,'MaxFunctionEvaluations',2000); 55 56% True or false to decide whether to run Genetic algorithm (Genetic 57% algorithm) 58stg.ga = false; 59 60% Options for Genetic algorithm (Genetic algorithm options) 61stg.ga_options = optimoptions(@ga,'MaxGenerations',200,... 62 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 63 'PopulationSize',stg.popsize,... 64 'MutationFcn','mutationadaptfeasible'); 65 66% True or false to decide whether to run Particle swarm (Particle swarm) 67stg.pswarm = false; 68 69% Options for Particle swarm (Particle swarm options) 70stg.pswarm_options = optimoptions('particleswarm',... 71 'MaxTime',stg.optt,'UseParallel',stg.optmc,'MaxIterations',200,... 72 'SwarmSize',stg.popsize); 73 74% True or false to decide whether to run Surrogate optimization (Surrogate 75% optimization) 76stg.sopt = false; 77 78% Options for Surrogate optimization (Surrogate optimization options) 79stg.sopt_options = optimoptions('surrogateopt',... 80 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 81 'MaxFunctionEvaluations',5000,... 82 'MinSampleDistance',0.2,'MinSurrogatePoints',32*2+1); 83endExample settings code
1% Time for the optimization in seconds (fmincon does not respect this 2% time!!) (Optimization time) 3stg.optt = 60*1; 4 5% Population size for the algorithms that use populations (Population size) 6stg.popsize = 10; 7 8% optimization start method, choose between: 1 Random starting point or 9% group of starting points inside the bounds 2 Random starting point or 10% group of starting points near the best point (Optimization start method) 11stg.osm = 1; 12 13% Distance from best parameter array to be used in stg.osm method 2 14% (Distance from best parameter array) 15stg.dbs = 0.1; 16 17% True or false to decide whether to use Multistart (Multistart) 18stg.mst = false; 19 20% Multistart size 21stg.msts = 1; 22 23% True or false to decide whether to display Plots (Plots doesn't work if 24% using multicore) (Optimization plots) 25stg.optplots = true; 26 27% True or false to decide whether to run fmincon (no gradient so this 28% doesn't work very well, no max time!!) 29stg.fmincon = false; 30 31% Options for fmincon (fmincon options) 32stg.fm_options = optimoptions('fmincon',... 33 'UseParallel',stg.optmc,... 34 'Algorithm','interior-point',... 35 'MaxIterations',2,'OptimalityTolerance',0,... 36 'StepTolerance',1e-6,'FiniteDifferenceType','central',... 37 'MaxFunctionEvaluations',10000); 38 39% True or false to decide whether to run simulated annealing (Simulated 40% annealing) 41stg.sa = false; 42 43% Options for simulated annealing (Simulated annealing options) 44stg.sa_options = optimoptions(@simulannealbnd, ... 45 'MaxTime',stg.optt,... 46 'InitialTemperature',... 47 ones(1,stg.parnum)*2,'ReannealInterval',40); 48 49% True or false to decide whether to run Pattern search (Pattern search) 50stg.psearch = false; 51 52% Options for Pattern search (Pattern search options) 53stg.psearch_options = optimoptions(@patternsearch,... 54 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 55 'UseCompletePoll',true,'UseCompleteSearch',true,... 56 'MaxMeshSize',2,'MaxFunctionEvaluations',2000); 57 58% True or false to decide whether to run Genetic algorithm (Genetic 59% algorithm) 60stg.ga = true; 61 62% Options for Genetic algorithm (Genetic algorithm options) 63stg.ga_options = optimoptions(@ga,'MaxGenerations',200,... 64 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 65 'PopulationSize',stg.popsize,... 66 'MutationFcn','mutationadaptfeasible','Display','diagnose'); 67 68% True or false to decide whether to run Particle swarm (Particle swarm) 69stg.pswarm = false; 70 71% Options for Particle swarm (Particle swarm options) 72stg.pswarm_options = optimoptions('particleswarm',... 73 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 74 'SwarmSize',stg.popsize); 75 76% True or false to decide whether to run Surrogate optimization (Surrogate 77% optimization) 78stg.sopt = false; 79 80% Options for Surrogate optimization (Surrogate optimization options) 81stg.sopt_options = optimoptions('surrogateopt',... 82 'MaxTime',stg.optt,'UseParallel',stg.optmc,... 83 'MaxFunctionEvaluations',5000,... 84 'MinSampleDistance',0.2,'MinSurrogatePoints',32*2+1); 85end
stg.optt - (double) Time for the optimization in seconds (fmincon does not respect this time!!)
stg.popsize - (double) Population size (for the algorithms that use populations)
stg.osm - (double) optimization start method, choose between
Get a random starting parameter set or group of starting parameter sets inside the bounds
Get a random starting parameter set or group of starting parameter sets near the best parameter set
stg.dbpa - (double) Distance from best parameter set to be used in stg.osm method 2
stg.mst - (logical) Decide whether to use one or multiple starting parameter sets for the optimization
stg.msts - (double) Number of starting parameter sets for the optimizations
stg.optplots - (logical) Decide whether to display optimiazation plots (They aren’t ploted if running the code in multicore)
stg.fmincon - (logical) Decide whether to run fmincon (no gradient in our models so this doesn’t work very well, does not respect time set for the optimization!!)
stg.fm_options - (optim.options.Fmincon) Options for fmincon
stg.sa - (logical) Decide whether to run simulated annealing
stg.sa_options - (optim.options.SimulannealbndOptions) Options for simulated annealing
stg.psearch - (logical) Decide whether to run Pattern search
stg.psearch_options - (optim.options.PatternsearchOptions) Options for Pattern search
stg.ga - (logical) Decide whether to run Genetic algorithm
stg.ga_options - (optim.options.GaOptions) Options for Genetic algorithm
stg.pswarm - (logical) Decide whether to run Particle swarm
stg.pswarm_options - (optim.options.Particleswarm) Options for Particle swarm
stg.sopt - (logical) Decide whether to run Surrogate optimization
stg.sopt_options - (optim.options.Surrogateopt) Options for Surrogate optimization
Automatically generated at Import¶
stg.expn - (double) Total number of experiments stored in the SBtab
stg.outn - (double) Total number of experimental outputs specified in the SBtab